Perturbation modeling with the SinkhornProblem#
In this tutorial, we showcase how to use the generic solver class :class:~moscot.problems.generic.SinkhornProblem to model cellular responses to chemical drugs.
Preliminaries#
import warnings
import moscot as mt
import moscot.plotting as mtp
from moscot.problems.generic import SinkhornProblem
from tqdm.std import TqdmWarning
import numpy as np
import matplotlib.pyplot as plt
import scanpy as sc
warnings.simplefilter("ignore", UserWarning)
warnings.simplefilter("ignore", TqdmWarning)
warnings.simplefilter("ignore", FutureWarning)
Dataset description#
The sciplex() dataset is a perturbation dataset published in [Srivatsan et al., 2020].
It contains transcriptomes of A549, K562, and mCF7 cells exposed to 188 compounds.
Data obtained from scPerturb.
adata = mt.datasets.sciplex()
adata
AnnData object with n_obs × n_vars = 799317 × 110984
obs: 'ncounts', 'well', 'plate', 'cell_line', 'replicate', 'time', 'dose_value', 'pathway_level_1', 'pathway_level_2', 'perturbation', 'target', 'pathway', 'dose_unit', 'celltype', 'disease', 'cancer', 'tissue_type', 'organism', 'perturbation_type', 'ngenes', 'percent_mito', 'percent_ribo', 'nperts', 'chembl-ID'
var: 'ensembl_id', 'ncounts', 'ncells'
Filter the data#
drugs = [
"Dacinostat (LAQ824)",
"Flavopiridol HCl",
"Givinostat (ITF2357)",
"TAK-901",
]
adata_red = adata[adata.obs["perturbation"].isin(["control"] + drugs)].copy()
adata_red
AnnData object with n_obs × n_vars = 29020 × 110984
obs: 'ncounts', 'well', 'plate', 'cell_line', 'replicate', 'time', 'dose_value', 'pathway_level_1', 'pathway_level_2', 'perturbation', 'target', 'pathway', 'dose_unit', 'celltype', 'disease', 'cancer', 'tissue_type', 'organism', 'perturbation_type', 'ngenes', 'percent_mito', 'percent_ribo', 'nperts', 'chembl-ID'
var: 'ensembl_id', 'ncounts', 'ncells'
sc.pp.normalize_total(adata_red, target_sum=1e4)
sc.pp.log1p(adata_red)
sc.pp.pca(adata_red)
sc.pp.neighbors(adata_red)
sc.tl.umap(adata_red)
sc.pl.umap(adata_red, color=["perturbation", "cell_line"], ncols=1)
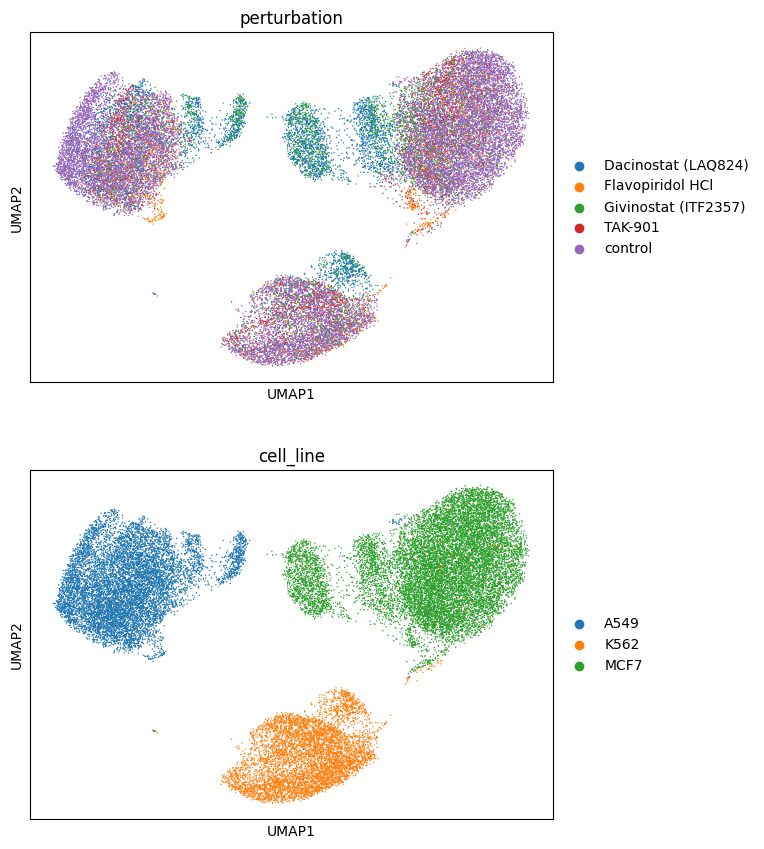
In this case, we want to compare the perturbations from the selected drugs with the control, so we use the star policy with the control as reference.
sp = SinkhornProblem(adata_red)
sp = sp.prepare(
key="perturbation", joint_attr="X_pca", policy="star", reference="control"
)
sp = sp.solve(1e-2, 0.95, 0.95)
sp
INFO Solving `4` problems
INFO Solving problem OTProblem[stage='prepared', shape=(3609, 17578)].
INFO Solving problem OTProblem[stage='prepared', shape=(3545, 17578)].
INFO Solving problem OTProblem[stage='prepared', shape=(2559, 17578)].
INFO Solving problem OTProblem[stage='prepared', shape=(1729, 17578)].
SinkhornProblem[('Dacinostat (LAQ824)', 'control'), ('Givinostat (ITF2357)', 'control'), ('TAK-901', 'control'), ('Flavopiridol HCl', 'control')]
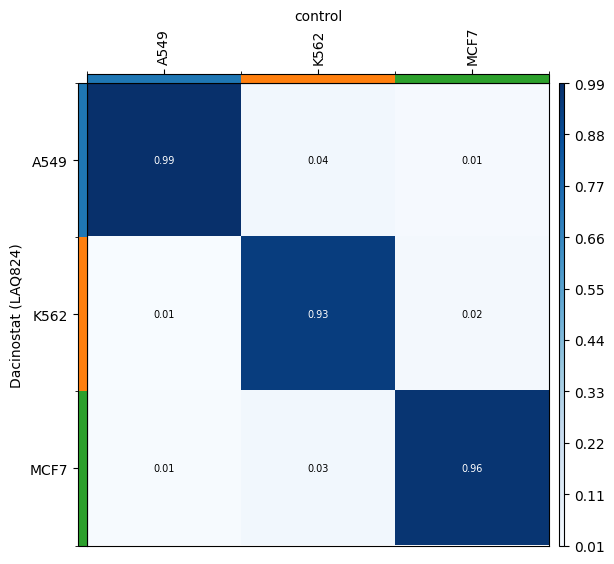
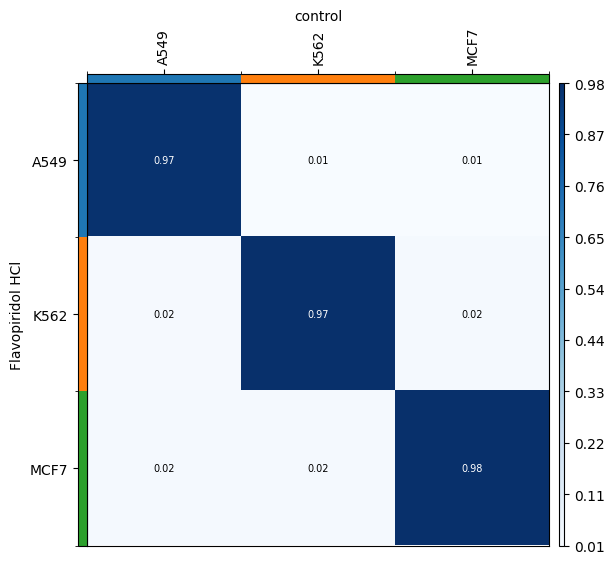
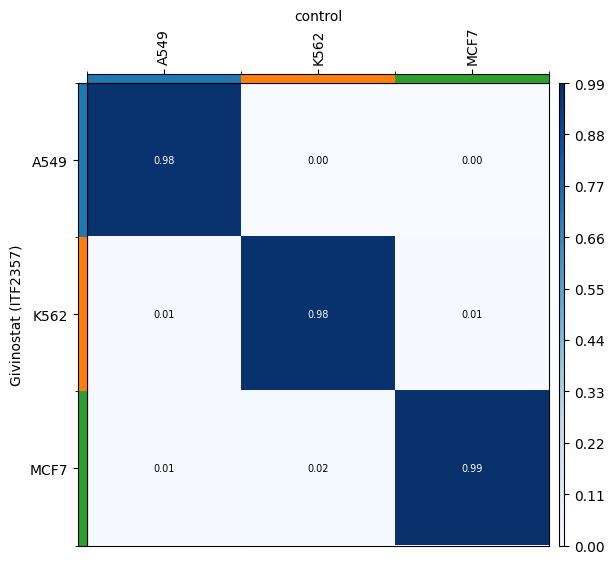
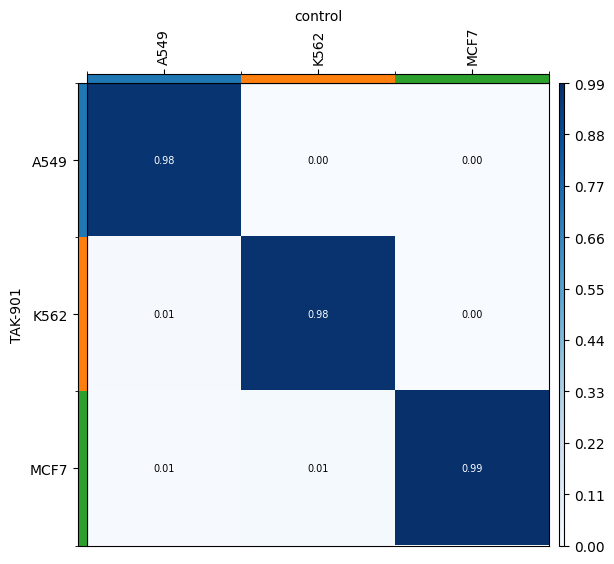
We can verify that the transport plan we learn is meaningful, as cell lines are mapped onto themselves.
for drug in drugs:
transition_matrix = sp.cell_transition(
drug, "control", "cell_line", "cell_line", key_added=f"cell_transition_{drug}"
)
mtp.cell_transition(sp, key=f"cell_transition_{drug}", cmap="Blues")
sc.tl.leiden(adata_red, key_added="new_clusters", resolution=0.9)
sc.pl.umap(adata_red, color=["new_clusters", "perturbation"], ncols=1)
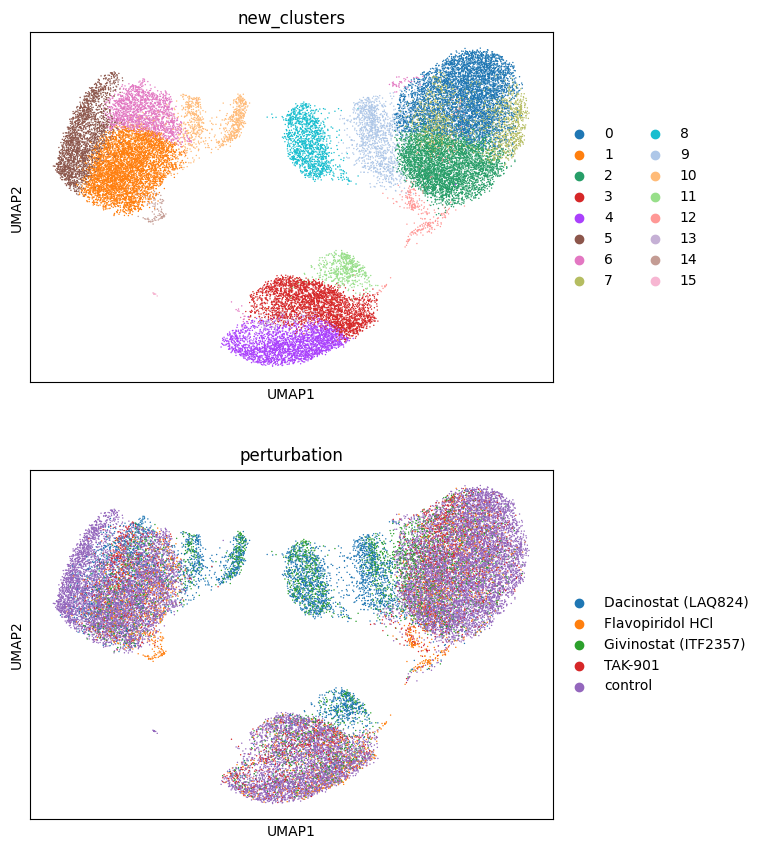
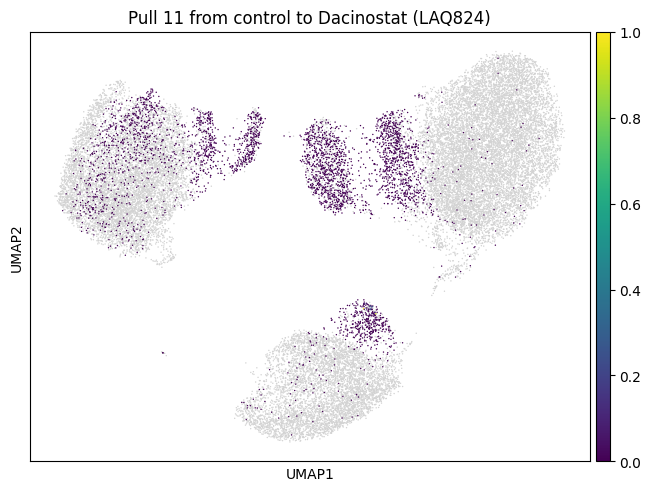
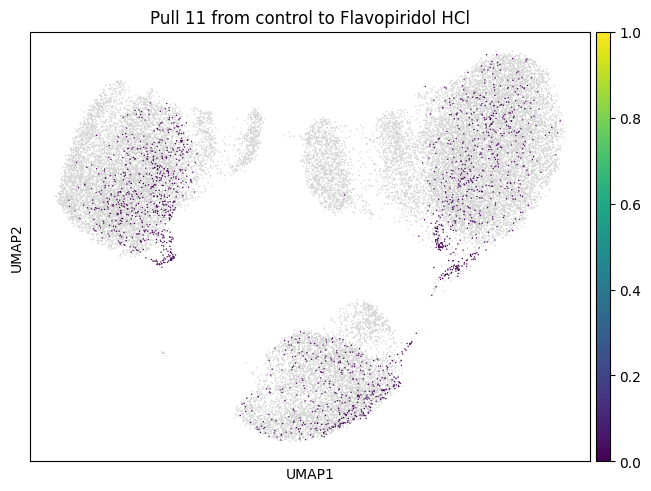
We now choose cluster 11 and see where it comes from
for drug in drugs:
sp.pull(
source=drug,
target="control",
data="new_clusters",
subset="11",
key_added=f"pull_1_{drug}",
)
for drug in drugs:
mtp.pull(sp, key=f"pull_1_{drug}")
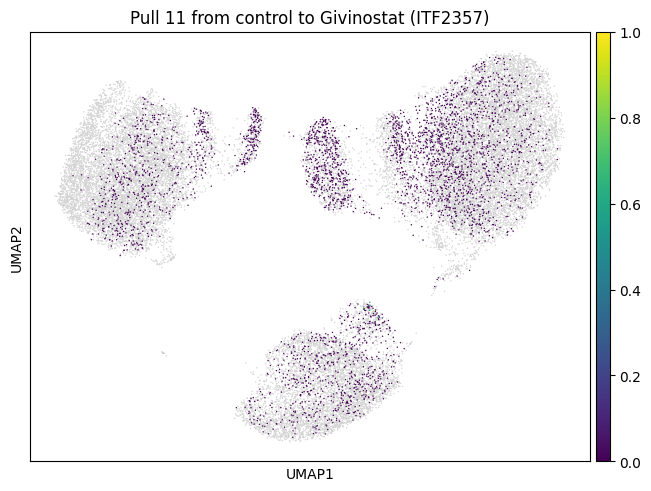
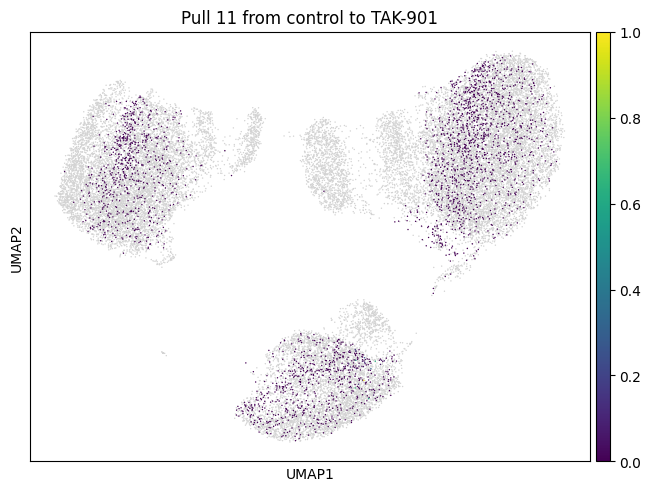
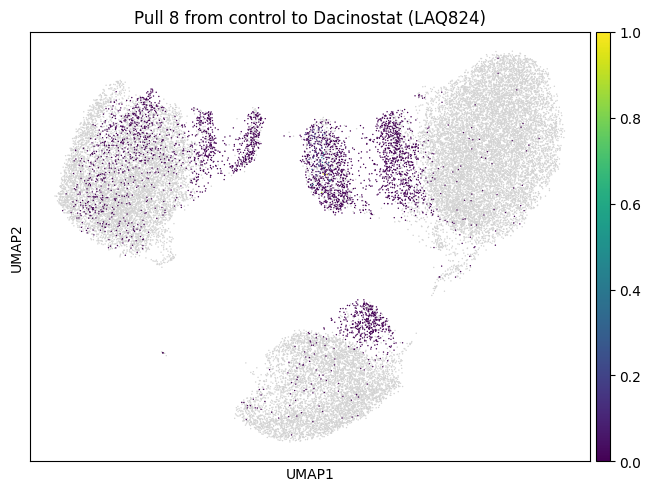
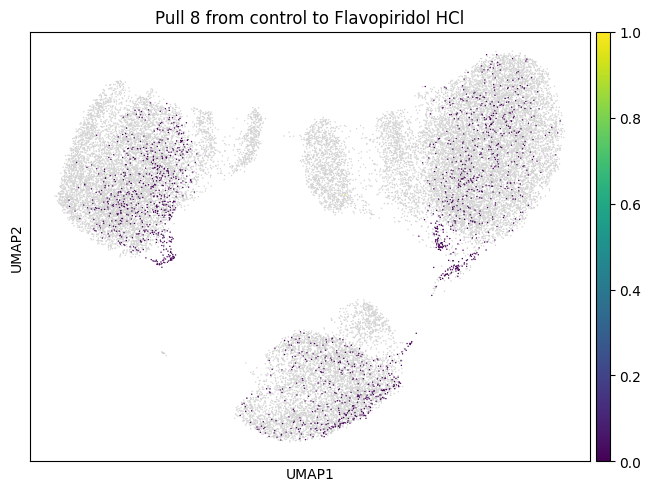
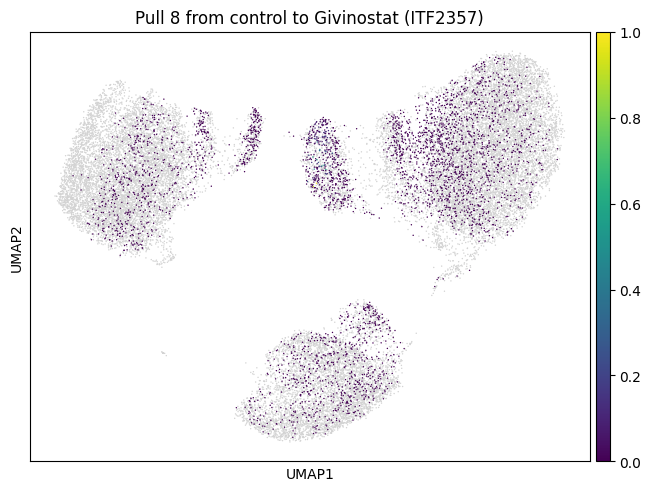
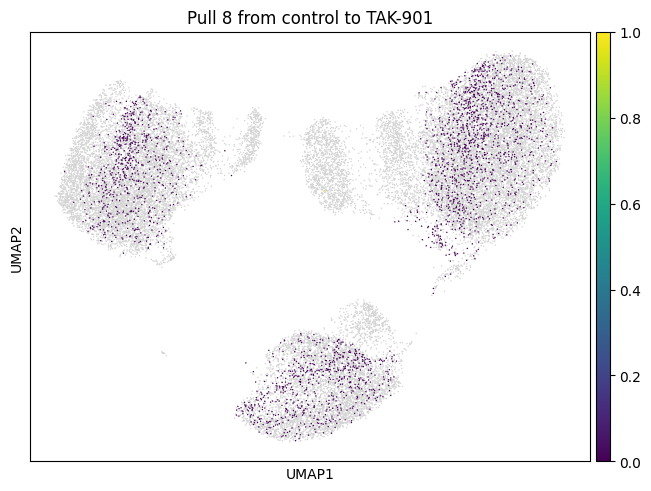
Let’s do the same for another cluster which is a bit bigger.
for drug in drugs:
sp.pull(
source=drug,
target="control",
data="new_clusters",
subset="8",
key_added=f"pull_2_{drug}",
)
for drug in drugs:
mtp.pull(sp, key=f"pull_2_{drug}")